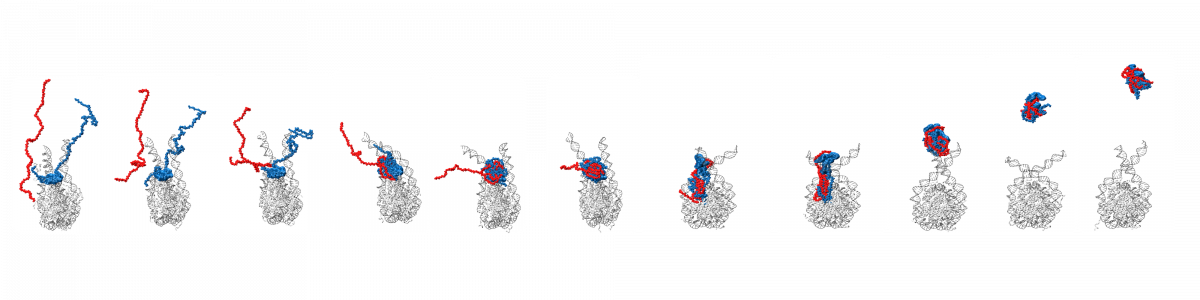
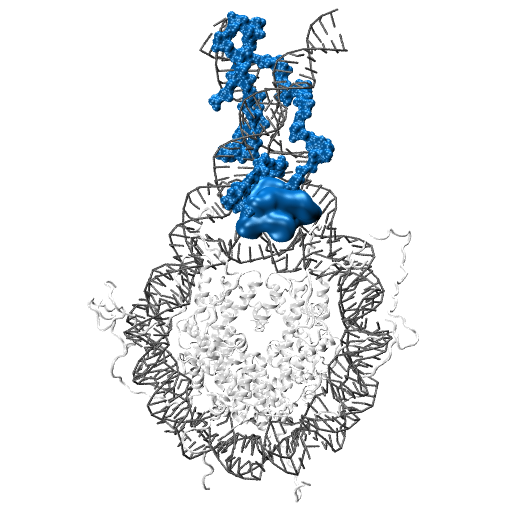
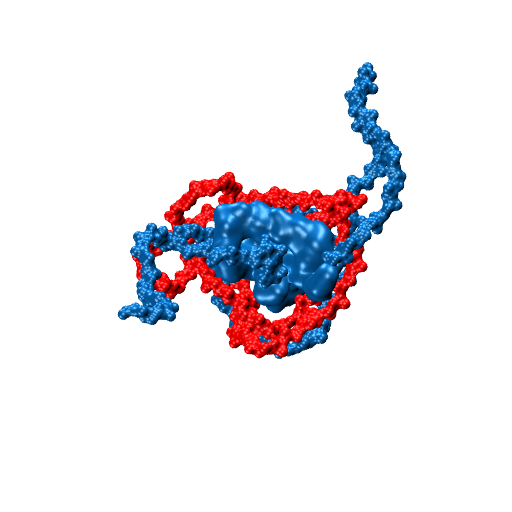
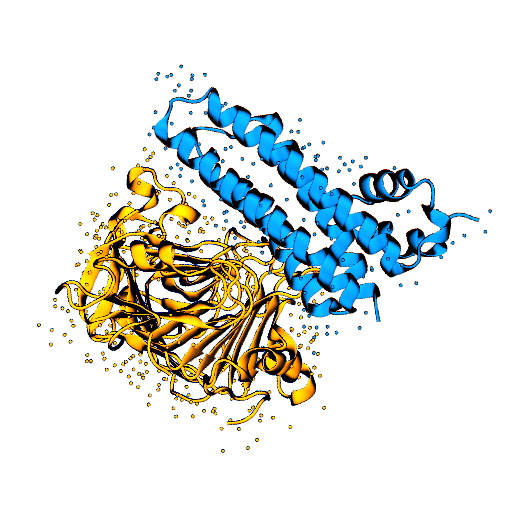
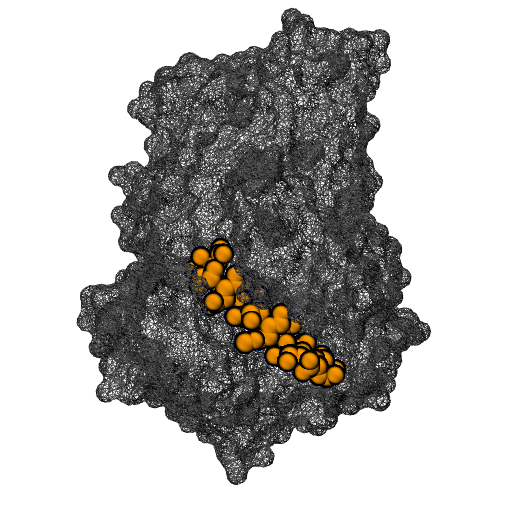
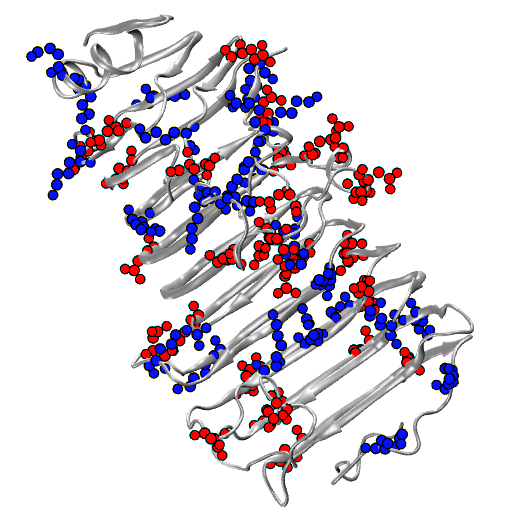
Molecular motors are able to convert chemical energy into mechanical energy or vice-versa. Other molecular motors are powered by an ion gradient, such as the rotary motor that synthesises ATP. Still other motors are powered by the free energy released when a nucleotide triphosphate is hydrolysed. The more processive a motor is the more unidirectional is the motion gleaned from the chemical free-energy released. In these motors, the free energy released constitutes a Brownian ratchet, preventing the reverse process. Remarkably, a class of enzymes called pectin methylesterase (PME) is involved in the processing of polysaccharide chains in plants, behaving as molecular motors that do not use high-energy exogenous co-substrates for their action, but carry the free energy for the Brownian ratchet endogenously. Understanding how PMEs achieve this, will lay the basis for the design of PME isoforms with enhanced or reduced processivity. Ultimately enhancing industrial processes and targeting events with serious economic downturns, such as plant parasitism and crop infection, in which PMEs play a big role.